The increased specificity for the BamHI-HF cut site has increased binding of the enzyme to the DNA. Restriction Enzymes are beleved to be evolved by bacteria to resist viral attack.

Bamhi An Overview Sciencedirect Topics
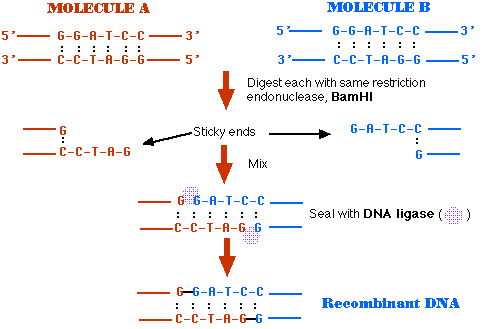
Recombinant Dna
1bhm Restriction Endonuclease Bamhi Complex With Dna
BamHI binds at the recognition sequence 5-GGATCC-3 and cleaves these sequences just after the 5.

Bamhi restriction endonuclease. The BamHI recognition site GGATCC was placed adjacent to a run of alternating GC base pairs in plasmid DNA. Single-buffer simplicity means more straightforward and streamlined sample processing. BamHI has a High Fidelity version BamHI-HF.
Restriction Enzyme Restriction Endonuclease Definition. Restriction enzymes are commonly classified into five types which differ in their structure and whether they cut their DNA. 240 County Road Ipswich MA 01938-2723 978-927-5054 Toll Free 1-800-632-5227 Fax.
High Fidelity HF Restriction Enzymes have 100 activity in rCutSmart Buffer. 1983 using the restriction endonuclease BamHI. The 73 sgRNAs targeting nonrepetitive sequence of MUC4 were cloned into the same sgRNA expression vector by amplifying the insertions using a common reverse primer but unique forward primers containing the.
Helixitta CC BY-SA 40. Also available as a FastDigest enzyme for rapid DNA digestion. Estas enzimas catalizan el corte de ADN doble banda en lugares específicos de forma reproducible y sin el uso de energía adicional.
Vào năm 1973 một nhóm các nhà khoa học đã tạo ra cơ thể sinh vật đầu tiên với các phân tử DNA tái tổ hợp. Theo đó Cohen ĐH Stanford Mỹ và Boyer ĐH California Mỹ cùng các cộng sự đã đưa được một đoạn DNA từ một plasmid này vào một plasmid khác tạo ra. To systematically map these pathways we developed a high-throughput screening approach called Repair-seq that measures the effects of thousands of genetic perturbations on mutations introduced at targeted DNA lesions.
For the library 2 the backbone plasmids were digested with SacII and BamHI the final products of 8177 bp. Restriction enzymes are Nucleases which can cleave the sugar-phosphate backbone of DNA found in bacteria. As they cut within the molecule they are commonly called restriction endonucleases.
Over 215 restriction enzymes are 100 active in a single buffer rCutSmart Buffer. This site was cut by the enzyme in relaxed DNA. A digestion of DNA by restriction endonuclease b denaturation of D N A on gel for hybridisation with specific probe c production of a group of genetically identical cells d isolation of DNA from a nucleated cell such as the one from the scene of crime 1 8 4.
Restriction enzymes are one class of the broader endonuclease group of enzymes. DNA sequence encoding the T7 RNA polymerase promoter upstream of the ShCas12k sgRNA was sourced as a gBlock IDT cloned into a pUC19 plasmid using restriction digest with. A vial of 6X Purple Load Dye is included with most restriction enzymes.
Restriction endonucleases cut the DNA double helix in very precise ways. 190 restriction enzymes are Time-Saver qualified meaning you can digest DNA in 5-15 minutes or digest DNA safely overnight. A restriction endonuclease that recognizes the sequence GGATC_C.
To perform restriction digestion of DNA with EcoR I and BamHI enzymes. Thermo Scientific BamHI restriction enzyme recognizes GGATCC sites and cuts best at 37C in its own unique buffer. BamHI-HF has the same specificity as BamHI NEB R0136 but it was engineered for reduced star activity.
La mayor parte requiere de la presencia de cofactores como el magnesio u otros cationes divalentes aunque algunas también requieren de ATP o S-adenosil metionina. Restriction enzyme also called restriction endonuclease is a protein produced by bacteria that cleaves DNA at specific sites along the molecule. See Reaction Conditions for Restriction Enzymes for a table of enzyme activity conditions for double digestion and heat inactivation for this and other restriction enzymes.
The validity of this approach was first demonstrated by Singleton et al. Then fragments 730 bp of error-prone PCR. Cells repair DNA double-strand breaks DSBs through a complex set of pathways critical for maintaining genomic integrity.
Under enzyme excess conditions the enzyme can. BamHI from Bacillus amyloliquefaciens is a type II restriction endonuclease having the capacity for recognizing short sequences 6 bp of DNA and specifically cleaving them at a target siteThis exhibit focuses on the structure-function relations of BamHI as described by Newman et al. The expression constructs for optimized sgRNAs were ordered as gBlocks from IDT and cloned into the lentiviral U6-based expression vector digested by XbaI and BamHI.
Restriction Endonuclease Restriction Enzyme is a bacterial enzyme that cuts dsDNA into fragments after recognizing specific nucleotide sequence known as recognition or restriction site. A restriction enzyme restriction endonuclease or restrictase is an enzyme that cleaves DNA into fragments at or near specific recognition sites within molecules known as restriction sites. A aa aaa aaaa aaacn aaah aaai aaas aab aabb aac aacc aace aachen aacom aacs aacsb aad aadvantage aae aaf aafp aag aah aai aaj aal aalborg aalib aaliyah aall aalto aam.
Introduction - Restriction Endonucleases are enzymes that produce internal cuts called cleavage In the DNA molecule. Esquema de reacción de la enzima de restricción HindIII Fuente.
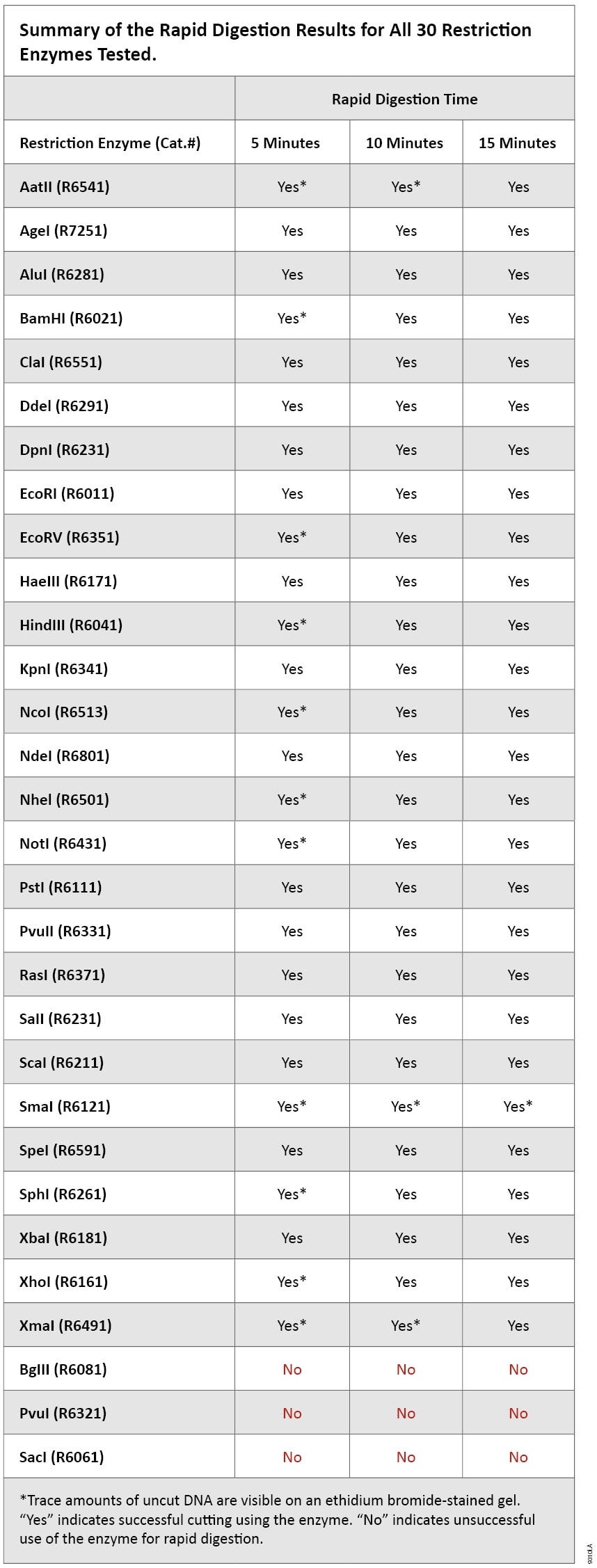
Rapid Dna Digestion Using Promega Restriction Enzymes
Bamhi Neb

Solved Q2 The Restriction Endonuclease Bamhi Recognizes Chegg Com

A Restriction Endonuclease Map Of The 9 Kb Bamhi Bamhi Genomic Download Scientific Diagram
3
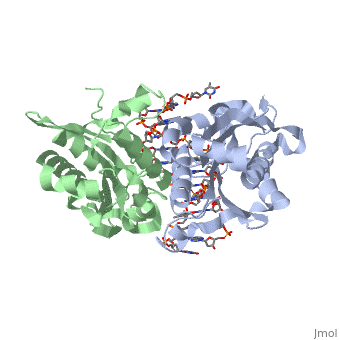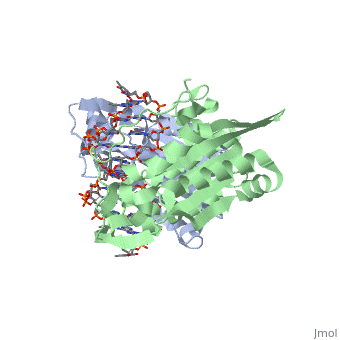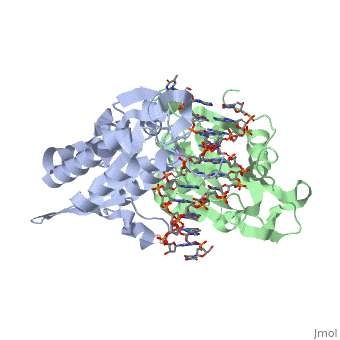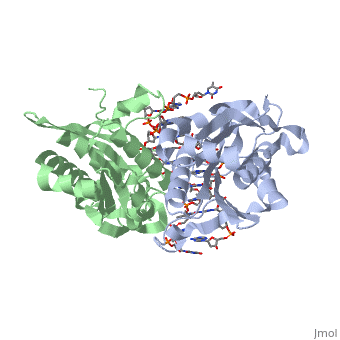
Bamhi Proteopedia Life In 3d
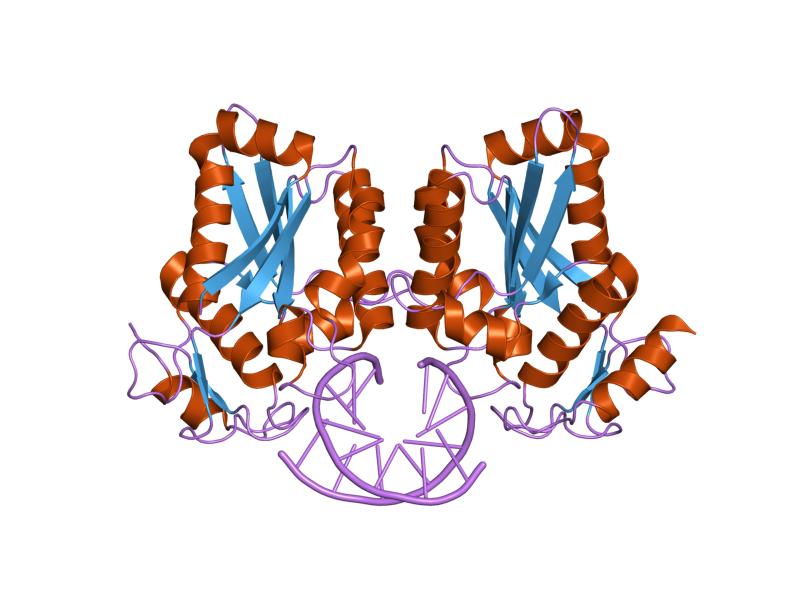
Bamhi Wikipedia

Bamhi Neb

